- Home
- size medium in numbers
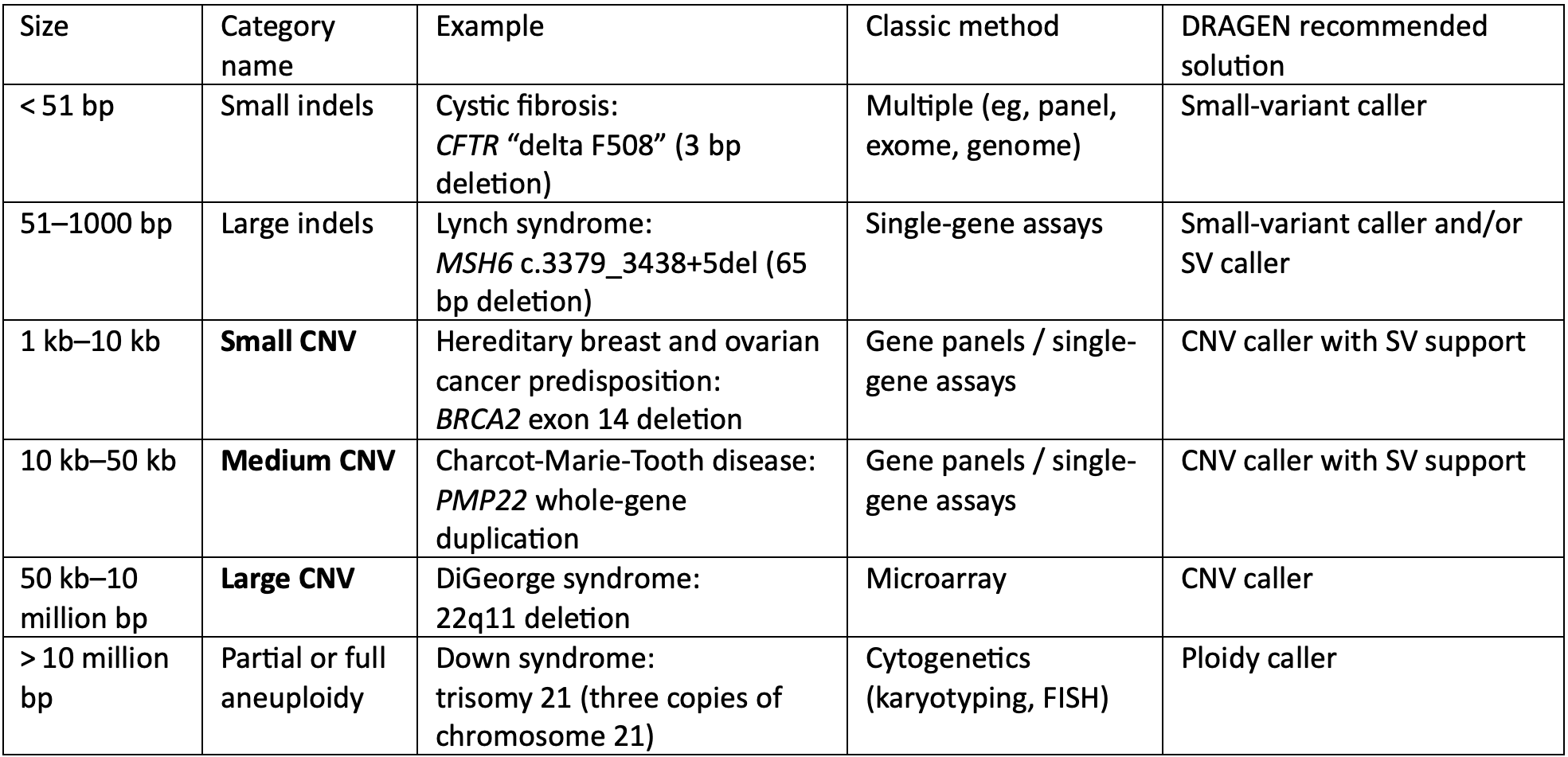
- Technical spotlight: Detecting small- and medium-length copy number variants by whole-genome sequencing
Technical spotlight: Detecting small- and medium-length copy number variants by whole-genome sequencing
4.5 (236) · $ 16.50 · In stock
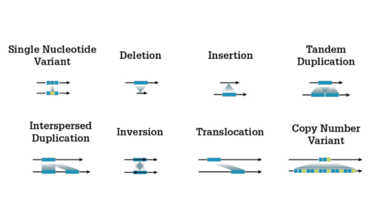
Historically, detecting different sizes of genetic variants has required using multiple different tests. By combining Illumina WGS with secondary analysis algorithms built into the DRAGEN Bio-IT Platform, researchers can achieve high-sensitivity detection of all these different variant types using a mixture of methods described here.
SMRT Long-Read Sequencing Solves Genetic Mysteries

Samuel Strom, PhD FACMG on LinkedIn: #womeninscience

Samuel Strom, PhD FACMG on LinkedIn: #genetics #acmg2023

Rami Mehio on LinkedIn: Building a resilient and scalable clinical
Single-cell copy number variant detection reveals the dynamics and diversity of adaptation
Copy Number Variation (CNV) Analysis

Rami Mehio على LinkedIn: Using whole-genome sequencing to evaluate

Rami Mehio on LinkedIn: GitHub - Illumina/DRAGMAP: DRAGEN open

The G Word Genomics England
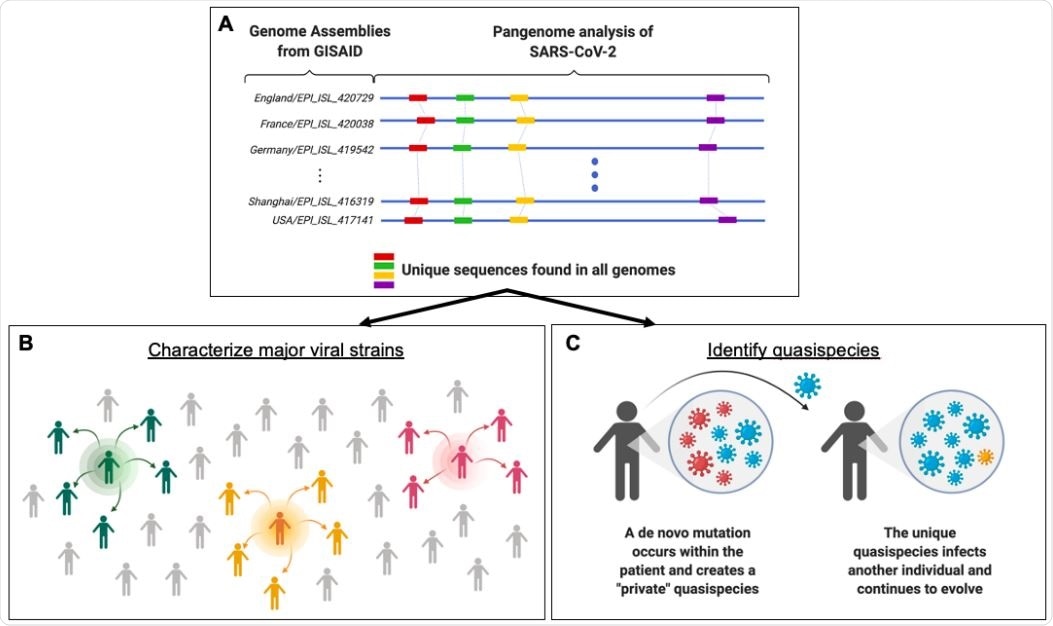
Study suggests SARS-CoV-2 mutation fingerprints could be used for molecular contact tracing
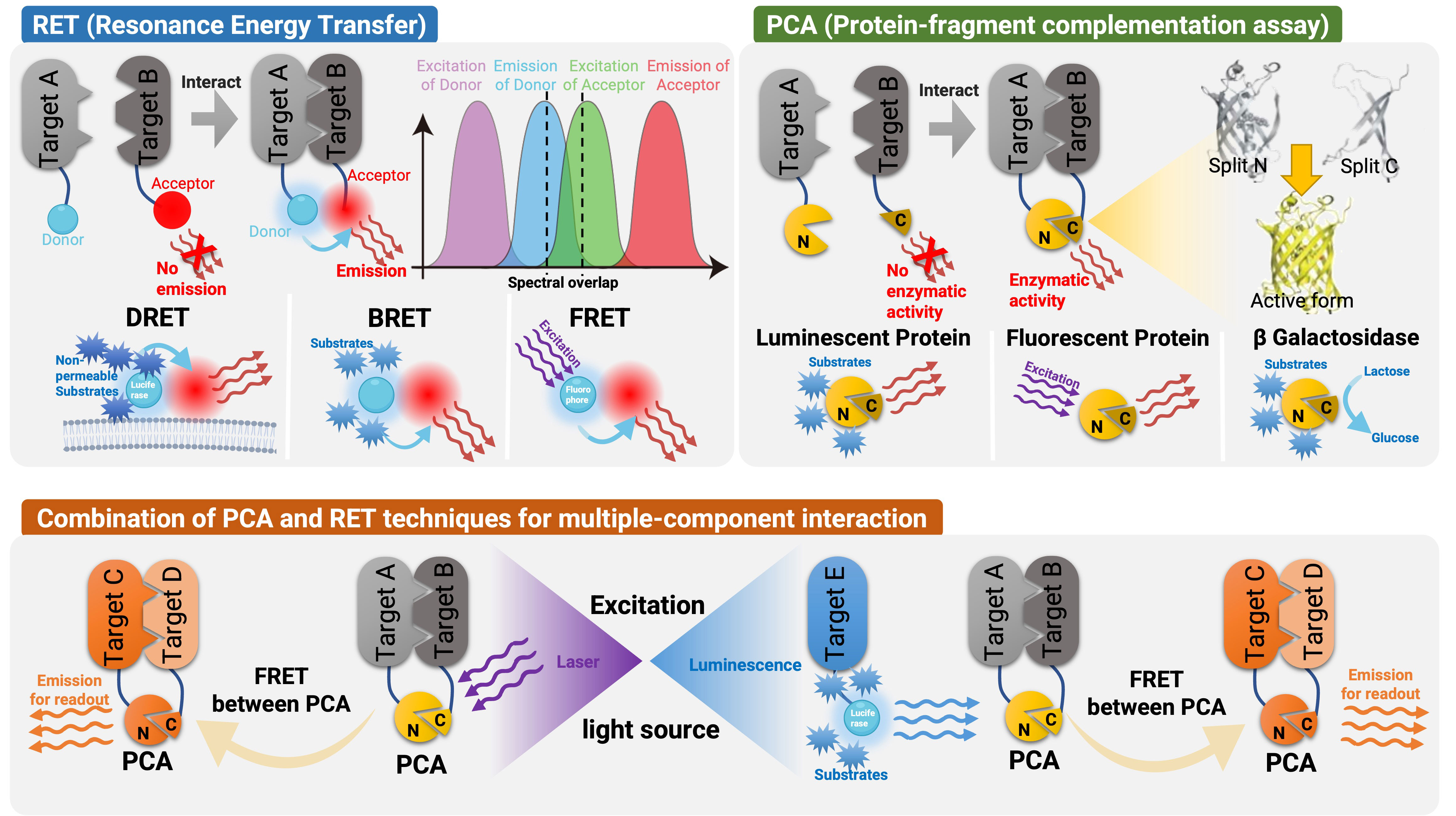
Frontiers Detecting and measuring of GPCR signaling – comparison of human induced pluripotent stem cells and immortal cell lines

Rosy Volpi on LinkedIn: Women in Science Day 2024

Copy number profiling using nanopore low-pass whole genome sequencing.

Rosy Volpi on LinkedIn: LinkedIn
Technical spotlight: Detecting small- and medium-length copy number variants by whole-genome sequencing









